티스토리 뷰
[과산화수소 반응성 단백질 표지 시약/H2O2-Responsive Protein Labeling Reagent] Hyp-Stamp [FDV-0052]_Funakoshi-코아사이언스
코피디 2024. 1. 31. 00:26H2O2-Responsive Protein Labeling Reagent
Hyp-Stamp
Tracer for H2O2-responsive proteins
Hyp-Stamp reacts with H2O2 and fluorescent labels proteins nearby H2O2
Cat.# FDV-0052 /Size: 100 µg
Hyp-Stamp is an H2O2-responsive protein labeling reagent.
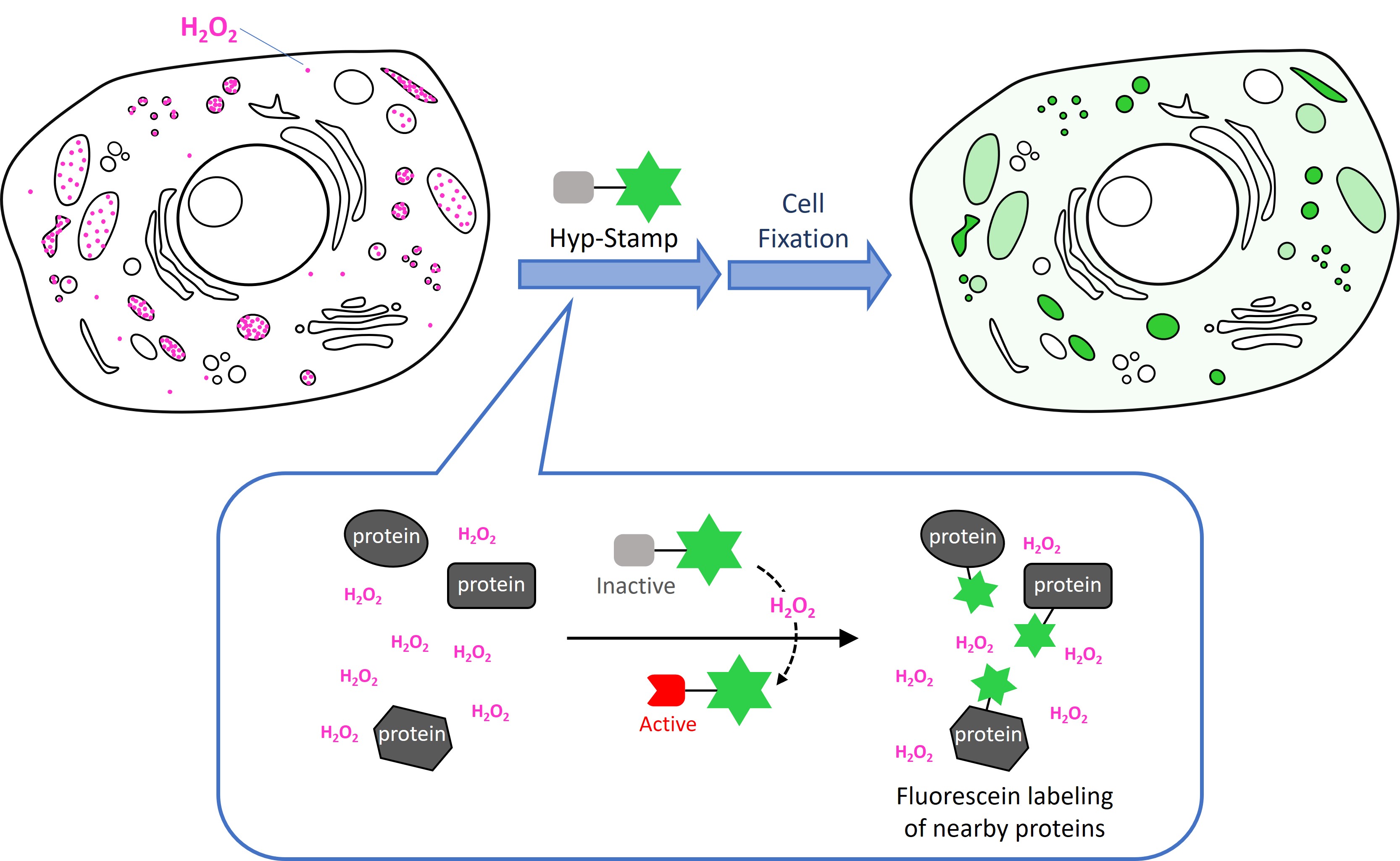
Hyp-Stamp responds to cellular H2O2 and accomplishes fluorescein labeling of proteins localized nearby H2O2. Since Hyp-Stamp-mediated fluorescein-labeled proteins remain at the original site, and after cell or tissue fixation, users can observe the H2O2 localization in fixed cells or fixed tissues by fluorescent microscope. Further, users can analyze and identify individual proteins nearby H2O2.
Overview of H2O2 monitoring with Hyp-Stamp
Product Background
ROS and H2O2 in cells
Reactive Oxygen Species (ROS) are reactive molecules produced in a wide range of biological processes. ROS are mainly produced during oxidative stress and damage lipids, proteins, and DNAs, which are concerned with cellular senescence and cause various diseases, such as cancer, inflammation, cardiovascular diseases, and neurodegenerative diseases. On the other hand, ROS are also generated in normal metabolic processes and play essential roles in cellular functions by regulating many signaling pathways. Hydrogen peroxide (H2O2 ) is one of the ROS and exhibits mild reactivity and chemical stability compared with other ROS. Therefore, H2O2 is an ideal cellular signaling molecule that can move and diffuse with sufficient distance. H2O2 regulates various physiological processes, including inflammation reactions and growth factor stimulation. For these reasons, investigating the H2O2 behavior in diverse types of cells has been an important topic in life science.
Conventional method to analyze cellular H2O2
Researchers conventionally use H2O2-responsive fluorescent probes that turns “ON” by reacting with H2O2 to analyze the cellular H2O2. Although they are valuable tools for observing H2O2 generation in living cells, they diffuse to other regions in the cell after reacting with H2O2, which makes it difficult to observe the exact localization of H2O2 generation over time. In addition, the fluorescent probes cannot be used for multistaining with antibodies because they are removed in cell fixation process.
Principle
Hyp-Stamp is a reagent developed by Prof. Hamachi and co-workers at Kyoto University. Hyp-Stamp consists of a protein labeling moiety and a Fluorescein moiety. Before reacting with H2O2, the protein labeling moiety is protected with boronic acid ester, which does not react to proteins in this “inactivate form”. The protein labeling portion converts to its “active form” (quinone methide form) by reacting with cellular H2O2, removing the protecting group. The “active form” moiety rapidly reacts with the nucleophilic groups of proteins that exist nearby cellular H2O2.
Structure of Hyp-Stamp and its reaction mechanism
Conventional H2O2-responsive fluorescent probes can observe H2O2 generation in living cells while they become undetectable in fixed cells because they are removed in the fixation process. On the other hand, Hyp-Stamp can visualize the local site of H2O2 after fixation of the cells, while it cannot observe H2O2 localization in living cells before fixation because of the overwhelmed fluorescence derived from the excess unreacted Hyp-Stamp.
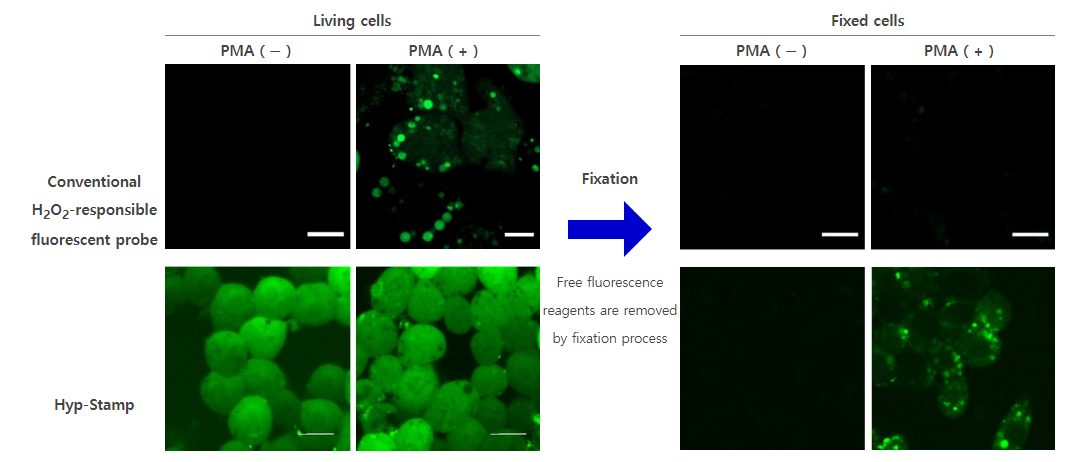
Fluorescent images of RAW264.7 cells in which H2O2 was generated by immune stimulation
Features
- Rapidly labels proteins nearby H2O2 with fluorescein.
- Reacts with H2O2 selectively compared to other major ROSs, such as superoxide (O2-), singlet oxygen (1O2), and hydroxy radical (·OH).
- Membrane permeable and taken up by cells spontaneously just by added in culture medium.
- Almost no cytotoxicity at the recommended concentration of 5 μM.
- Localizations of fluorescein-labeled proteins can be analyzed with fluorescent microscopy after cell fixation. Multistaining of the fluorescein-labeled proteins with antibodies for the protein of user interest is also compatible.
- Fluorescein-labeled proteins are applicable to detection or immunoprecipitation with anti-fluorescein antibody.
- Formulation: C41H38BFN2O11
- Molecular weight: 764.56 g/mol
- Solubility: DMSO
- Ex/Em: 495 nm/515 nm (FITC filter sets are available.)
Application data
Application classification
Application 1 : Multistaining of the fluorescein-labeled proteins with antibodies in immunostimulated macrophage cells
RAW264.7 cells were immunostimulated (treated with 1 ug/mL PMA) and incubated for 30 minutes. Afterward, 5 uM Hyp-Stamp was added, and the cells were cultured for 30 minutes. Then, cells were fixed with cold methanol or 4% formaldehyde and immunostained with mitochondrial proteins Sdha and Hspa9. Confocal laser scanning microscope imaging shows the immunofluorescence against Sdha and Hspap partially merged with fluorescein-labeled vesicles.
Application 2 : Imaging of the H2O2 localization in mouse hippocampal slices
Murine hippocampal slices were treated with mitochondrial Complex II inhibitor TTFA (1 mM, 30 min) to induce oxidative stress, then 5 uM Hyp-Stamp was added and incubated for 1 hour. The slices were fixed with 4% formaldehyde and permeabilized with cold methanol, followed by fluorescent imaging with a confocal laser scanning microscope. Localization of TTFA-triggered H2O2 was observed in the hippocampal slices.
Application 3 : Analysis of total fluorescein-labeled proteins under oxidative stress of mitochondria
HeLa cells were treated with mitochondrial Complex II inhibitor TTFA (1 mM, 30 min) to induce oxidative stress, then 5 uM Hyp-Stamp was added and incubated for 30 min. After preparing a cell lysate, samples are applied to an SDS-PAGE, imaged by Coomassie Brilliant Blue (CBB), and in-gel fluorescence. Results revealed the amount of fluorescein-labeled protein increased by TTFA treatment, while TTFA did not affect the amount of total protein.
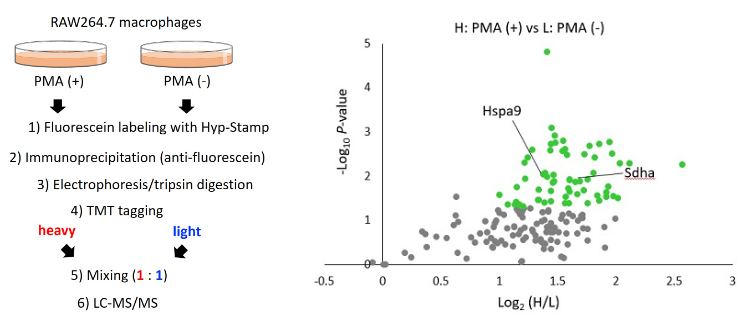
Application 4 : Identification of proteins existing nearby H2O2 generating site by proteomics methodology.
5 uM Hyp-Stamp was added to RAW264.7 cells in which immunostimulation (treated with 1 ug/mL PMA for 30 minutes) was given (+) or not (-), and the cells were cultured for 30 minutes. After preparing the cell lysates, immunoprecipitation was performed with an anti-fluorescein antibody to enrich fluorescein-labeled proteins. Proteins obtained by immunoprecipitation were applied to in-gel trypsin digestion to produce peptide solutions. After modifying the PMA(+) and PMA(-) samples with TMT heavy tag and TMT light tag, respectively, the two samples were mixed in a 1:1 ratio and subjected to LC-MS/MS analysis to investigate changes between samples comprehensively. Figures show the experimental scheme of proteomics analysis (left) and volcano plot of protein signal changes by PMA(+)/(-) (right). In the plot, proteins with log2(PMA added/not added) > 1, and P value < 0.05 are shown in green dots. Mitochondrial proteins such as Sdha and many vesicle proteins were identified, suggesting that these proteins exited nearby H2O2 generated by immunostimulation.
Reference
- Zhu, H., et al., "Imaging and profiling of proteins under oxidative conditions in cells and tissues by hydrogen-peroxide-responsive labeling.", J. Am. Chem Soc., 142(37), 15711~15721 (2020). [PMID:32822179]
추천 상품
coresciences 코아사이언스 Funakoshi co., ltd. 후나코시 한국 공식대리점 일본 후나코시
'실험.연구용 추천상품' 카테고리의 다른 글
- Total
- Today
- Yesterday
- gradient gel
- 코아사이언스
- OLED
- 건스터바이오텍
- 면역화학분석
- 오토클레이브백
- Coresciences
- matrix-driven delivery pellet
- filter
- 연속절편
- Sterlitech
- 형광염색
- 조직절편
- material science
- 후나코시
- Funakoshi
- 파라핀 블록
- 바이오헤저드백
- time release pellets
- 콘드로이틴 황산 올리고당
- 바이오하자드백
- solar cells
- 조직절편제작
- 고수용성 콘드로이틴
- 막필터
- 미니멸균백
- OPV
- 조직염색절편제작
- 파라핀 블럭
- allview PAGE buffer
| 일 | 월 | 화 | 수 | 목 | 금 | 토 |
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | ||
| 6 | 7 | 8 | 9 | 10 | 11 | 12 |
| 13 | 14 | 15 | 16 | 17 | 18 | 19 |
| 20 | 21 | 22 | 23 | 24 | 25 | 26 |
| 27 | 28 | 29 | 30 |